Week 5: Genotyping, Gels, GSEA, and Gene Regulatory Network Analysis!
March 29, 2024
Guten Morgen allerseits and hello from Switzerland! This week I left a little early due to spring break travels to Europe, but I was still able to make progress on my project though and learn so much from my mentor!
Last Friday, I got to perform another genotyping experiment! Over the weekend, I observed my lab advisor perform an RNA extraction (which was perhaps one of the most careful and rigorous experiments to be performed). The RNA extraction actually starts with DNA from the somatic cells which are bust open to reveal the CARLIN-edited DNA (DNA from the cells of CARLIN mice). CARLIN mice are the CRISPR array repair lineage tracing (CARLIN) engineered mouse line. The CARLIN system uses CRISPR-Cas9 to induce genetic modifications to generate cell barcodes that we can track and read out later for lineage tracing. Lineage tracing involves labeling cells at a particular time in development and then tracking them as they progress to see their differentiation and the cells/tissues they gave rise to (we can map cell fate and cell clones to the cells they came from).
The DNA from the CARLIN-edited mice is transcribed into RNA to produce mRNAs (per the central dogma). In an RNA extraction experiment, the mRNAs are transcribed back into single-stranded cDNA (complementary DNA) using reverse transcriptase. Since the cDNAs were transcribed from functional, full processed mRNAs, they don’t contain the introns, or the noncoding regions of DNA. Looking at the cDNAs, we can now see the parts of the genome that actually contained the CARLIN barcodes!
On Monday, I watched my advisor plug-check and eartag mice again (eartagging just involved putting a numbered tag in a mouse’s ear so we have some way of identifying them!) while I helped them plan crosses and set up breeding cages. The rest of the day was pretty much all spent in coding! I’ve been performing Gene Set Enrichment Analysis (GSEA) and generating GSEA plots on the results from our differential gene expression (DGE) analysis on our single-cell RNA sequencing dataset. Essentially, we are taking annotated gene sets and pathway lists and trying to see how much of those genes and pathways are represented in our data, specifically in our list of genes that our DGE analysis ranked by differential expression metrics. We hope to identify how much of the genes in the gene sets that we pull are expressed by the cells we are analyzing, using statistical thresholds to understand enrichment significance.
I made a series of barplots and dotplots to visualize this enrichment! Here are a couple examples:

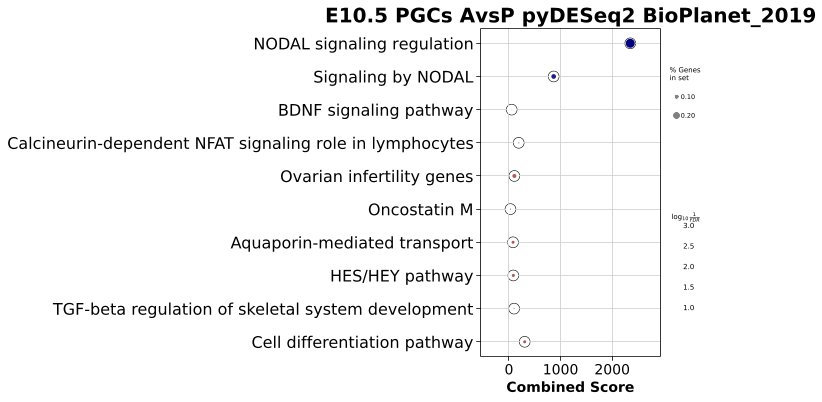
On Wednesday, I ran another genotyping experiment and since then, I’ve been working on pySCENIC analysis with my advisor. Python Single-Cell Regulatory Network Inference and Clustering (pySCENIC) involves checking the cluster of genes (known as regulons) that a list of transcription factors regulate. Knowing how and which transcription factors control gene expression helps us understand gene regulatory networks and interactions, and more broadly, and as a result, heterogeneity among the migrating PGCs.
Finally, I got to try my hand at zebrafish embryo dissections! (P.S. they’re very hard.)
Auf Wiedersehen! That’s it for me! Super duper thankful again for all the learning I get to do and for my mentor’s support and guidance — more on all the things I get to learn and do next time!

Leave a Reply
You must be logged in to post a comment.