Week 7: Genotyping, Imaging, and Sorting!
April 19, 2024
Hi everyone!!
This week, I wasn’t able to do much coding at all, mostly because I was trying to get the R environment for CellChat — an “R package designed for inference, analysis, and visualization of cell-cell communication from single-cell and spatially resolved transcriptomics” — working. Unfortunately, it has not been working! However, I will be meeting today with my advisor to resolve that.
On Monday, my advisor and I did another FACS sort to get cells to make a few more replicates of our reconstituted embryonic mouse ovaries. P.S. My dissection skills have significantly improved! I’m able to dissect out the embryos to retrieve nearly all the gonads of the male embryos a lot quicker than before! As you might be able to tell, embryonic mouse ovaries are incredibly tiny, which means to observe and properly document their growth and the proliferation of the cells, we must do a lot of imaging. Monday, Tuesday, and Thursday, I imaged the ovaries using different lasers under the microscope and under different magnifications. So far, they look good! We also transferred the ovaries from the 96 well plate to the transwell after 4 days and made sure to change media in between as they progressed, more of which will be done early next week!
Images time!
Here is a picture of the 1k, 3k, and 5k replicates, all at 10x magnification and under the brightfield laser. These were on the 3rd day of being in the transwell, so they were in their 7th day of culture.
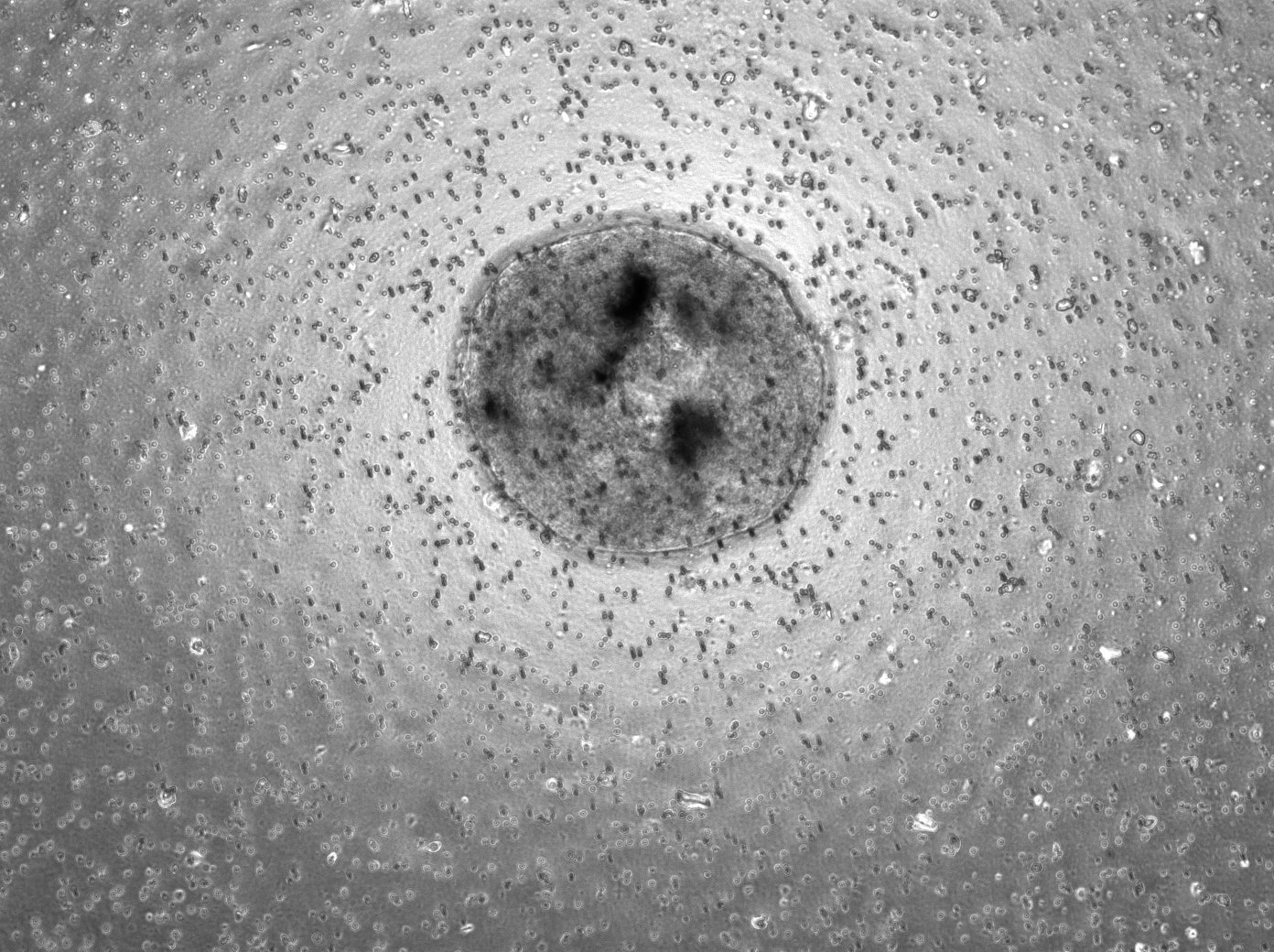

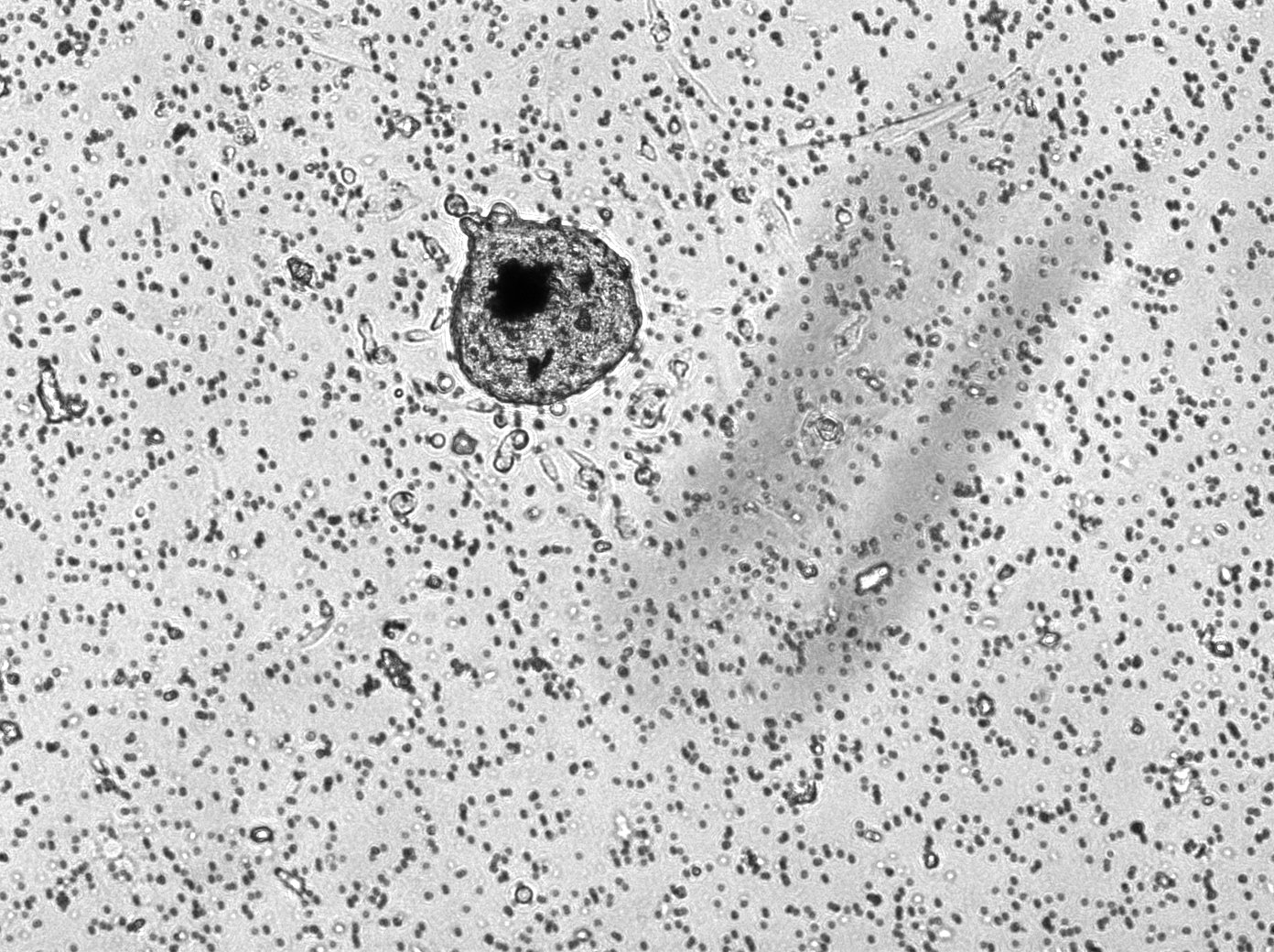
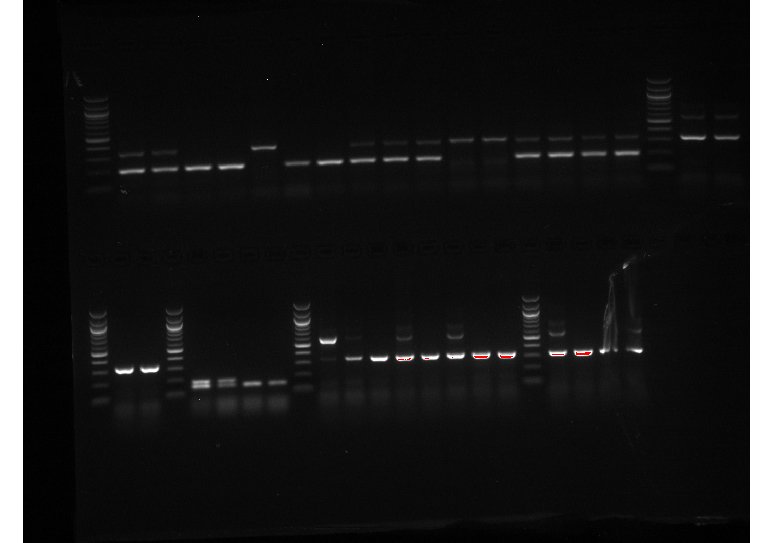
In addition to a lab meeting on Wednesday (and a very interesting paper discussion!), I mostly spent this week doing more genotyping and reconstituted ovary imaging (in addition to one more FACS on Thursday). There were a lot of new mice litters, so a lot of genotyping to do! I’ve attached a picture of one of the genotyping experiments I ran this week!
I also got the opportunity to watch my advisor do a library prep, where they took cDNA (generated from an RNA extraction – see Week 5’s post for more information on RNA extraction and genotyping), tagged it with UMIs (unique molecular identifiers), used size selection to ensure that only the cDNAs and not the other non-reverse transcribed RNAs were selected, and then performed PCR to extend the cDNAs. After all of this, they ensured that there were no cDNA duplicates (that the cDNAs that remain have unique UMIs). These cDNAs will then be sent to a sequencing lab, which will tell us the genetic composition of the cDNAs and where in our genome the genetic barcodes that the mice embryos have are!
More updates next week. Thank you for joining me! See you soon.

Leave a Reply
You must be logged in to post a comment.